Creating algorithms#
A central concept of Imaging Server Kit is the idea of converting a Python function into an “algorithm” object. Server Kit algorithms can be turned into web servers, widgets in Napari, and more.
In this section, we will cover the basics of how to create Server Kit algorithms.
Prerequisites#
Before continuing, make sure to have installed the required packages:
!pip install imaging-server-kit napari-serverkit
Note
You can download the code from this section as a Jupyter notebook from the download button at the top of this page (select .ipynb) to execute it locally on your computer.
Overview#
Creating an algorithm typically involves the following steps:
Write your image processing logic as a standard Python function.
Decorate it with
@sk.algorithm.Annotate parameters to control how they are interpreted and validated.
Annotate return values so that outputs are correctly displayed and managed.
Step 1: Define a Python Function#
Your processing logic should be encapsulated in a Python function. For example, here is a Gaussian filter:
from skimage.filters import gaussian
def gaussian_algo(image, sigma=1.0):
filtered = gaussian(image, sigma=sigma, preserve_range=True)
return filtered
This function takes an input image (as a NumPy array), applies a Gaussian filter to it, and returns another array containing the filtered image. The intensity of the filter is controlled by the value of the sigma parameter.
Step 2: Turn the Function into an Algorithm#
To convert this function into to an algorithm, import the imaging-server-kit package and use the @sk.algorithm decorator:
from skimage.filters import gaussian
import imaging_server_kit as sk
@sk.algorithm
def gaussian_algo(image, sigma=1.0):
filtered = gaussian(image, sigma=sigma, preserve_range=True)
return filtered
With your function wrapped with @sk.algorithm, it becomes an algorithm object.
type(gaussian_algo) # <imaging_server_kit.core.algorithm.Algorithm>
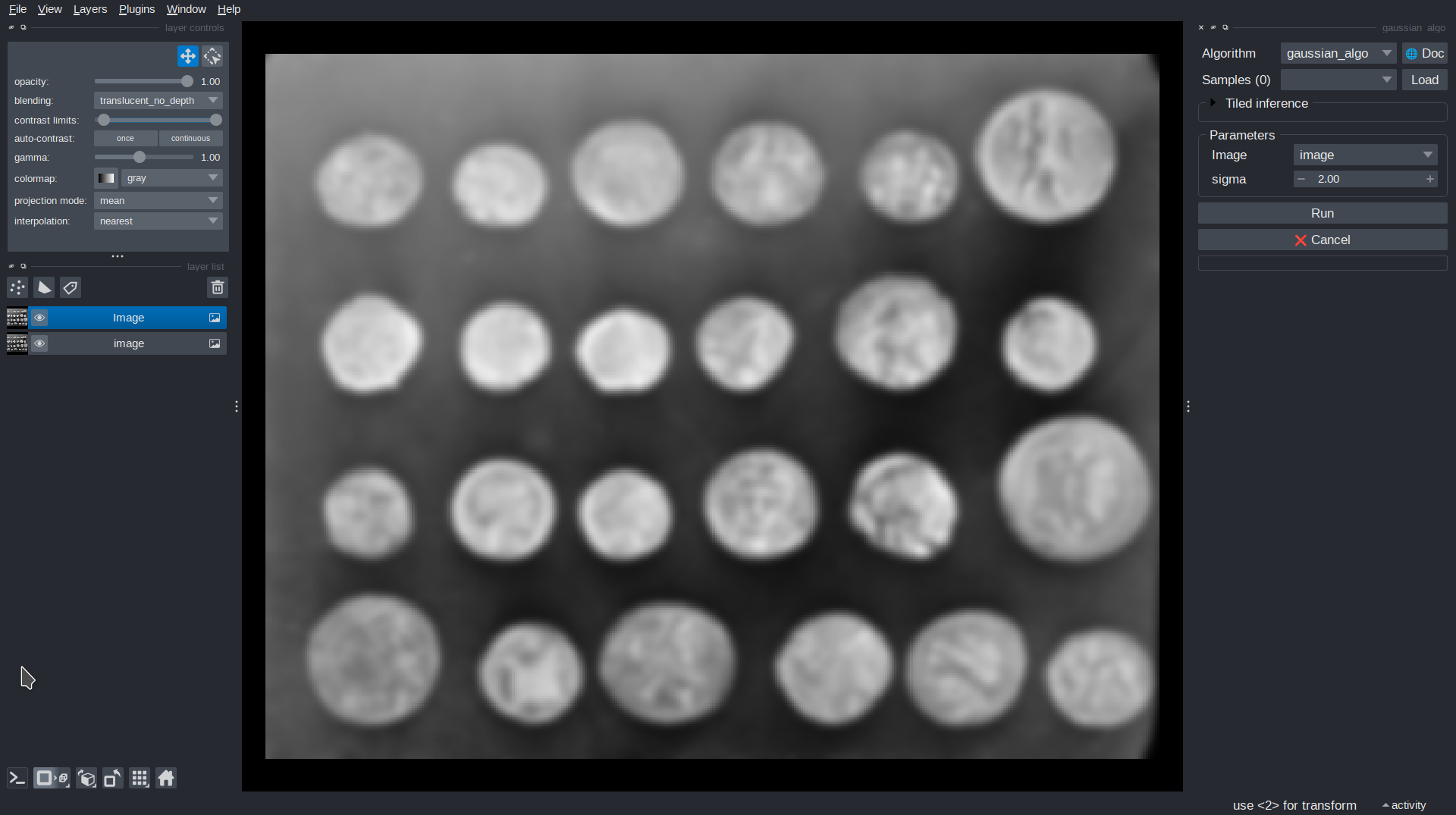
You can run and test it in Napari using:
viewer = sk.to_napari(gaussian_algo)
You can also add an example image to the viewer, for testing:
import skimage.data
image = skimage.data.coins()
viewer.add_image(image)
A Napari viewer should open, with your algorithm available as a dock widget. You can and apply the filter to the input image and adjust sigma to control the intensity of the blur.
Note
sk.to_napari() will only be available if you have installed the napari-serverkit plugin.
At this stage, the algorithm works, however a few things are not optimal. For example, users could enter a negative value for sigma, which would raise an error. To fix this, we need to annotate parameters.
Step 3: Annotate Parameters#
Annotating parameters tells Imaging Server Kit how to interpret parameters (the function arguments). In the function-to-algorithm conversion, each parameter is matched with a data layer, which allows it to be correctly validated and displayed in user interfaces.
Here is an improved version of our Gaussian filter algorithm, where we’ve annotated the sigma parameter via parameters={} inside the @sk.algorithm decorator, giving it a minimum and a default value:
@sk.algorithm(parameters={"sigma": sk.Float(min=0, default=1.0)})
def gaussian_algo(image, sigma):
filtered = gaussian(image, sigma=sigma, preserve_range=True)
return filtered
If you run this algorithm in Napari, you will see sigma can no longer be set to a negative value.
Data layers
Data layers are a key concept in Imaging Server Kit. They are used to store data, as well as to describe the meaning of input parameters and algorithm outputs.
In the example above, sk.Float is a data layer representing a floating-point value. It supports constraints such as:
min,max, anddefaultstep(for UI sliders)name(label next to the parameter)
There are many more data layers available. For a full list, see: Data layers
While parameters={} is the most explicit and reliable annotation method, you can also use:
Type hints:
int,float,bool,str, the data layer types (sk.Float…) ornp.ndarray- interpreted as an image type.Default values: the type of the default value is inferred as the parameter type.
Variable names: the variable names
image,mask,points, etc., are automatically inferred using the corresponding data layer.
A combination of these methods is also possible. We use the following priority order for the interpretation:
Explicit annotations (
parameters={})Type hints
Default values
Variable names
For example, the following parameter annotations are all valid:
# Explicit annotation
@sk.algorithm(parameters={"img": sk.Image(), "sigma": sk.Float()})
def gaussian_algo(img, sigma):
...
@sk.algorithm
def gaussian_algo(image, sigma=1.0): # <- `image` variable name, default sigma
...
@sk.algorithm
def gaussian_algo(img: sk.Image, sigma: float): # <- Type hints
...
Step 4: Annotate return values#
Similarly to parameters, return values should be annotated to specify what they represent. This allows Imaging Server Kit to assign each return value to a data layer and to correctly interpret and display it in Napari or QuPath.
For example, let’s take a look at a simple threshold algorithm:
@sk.algorithm(
name="Intensity threshold", # <- You can also give a name to the algorithm
parameters={
"threshold": sk.Integer(name="Threshold", min=0, max=255, default=128, auto_call=True)
}
)
def threshold_algo(image, threshold):
mask = image > threshold
return sk.Mask(mask) # <- Return the binary mask array as a `sk.Mask`
In this example, the algorithm applies an intensity threshold to segment the input image. We have annotated the threshold parameter to restrict it to values betwen 0 and 255. Moreover, we’ve used auto_call=True to specify that the algorithm automatically re-runs when the threshold changes in the UI.
As for the returned binary array, we would like it to be interpreted as a segmentation mask. Therefore, we have wrapped it into a sk.Mask data layer. As a result, when running this algorithm in Napari, the binary mask is correctly displayed as a Labels layer. In QuPath, it would be represented as a polygon annotation.
Complete example#
A more complete version of a segmentation algorithm combining a Gaussian filter with an intensity threshold might look like this:
import imaging_server_kit as sk
from skimage.filters import gaussian
@sk.algorithm(
name="Segmentation pipeline",
description="A simple pipeline for segmenting images.",
parameters={
"image": sk.Image(),
"sigma": sk.Float(
name="Sigma",
description="Intensity of the blur applied before thresholding the image.",
min=0,
default=1.0,
step=0.5,
auto_call=True,
),
"threshold": sk.Float(
name="Threshold (rel.)",
description="Intensity threshold, relative to the image min() and max() values.",
default=0.5, min=0, max=1, step=0.1,
auto_call=True,
),
"dark_background": sk.Bool(name="Dark background", default=True, auto_call=True)
},
)
def threshold_algo(image, sigma, threshold, dark_background):
# Apply a Gaussian filter
blurred_image = gaussian(image, sigma=sigma, preserve_range=True)
# Compute the absolute threshold to apply
thresh_abs = threshold * (blurred_image.max() - blurred_image.min())
# Binarize the image
if dark_background:
mask = blurred_image > thresh_abs
else:
mask = blurred_image <= thresh_abs
# Compute the area fraction of the mask
fract = mask.sum() / mask.size
# Return all annotated outputs
return (
sk.Image(blurred_image, name="Blurred", meta={"colormap": "viridis"}),
sk.Mask(mask, name="Binary mask"),
sk.String(f"Area fraction: {fract:.02f}", name="Fraction of True pixels"),
)
sk.to_napari(threshold_algo)
Notice that this algorithm
produces several outputs of different types:
sk.Image,sk.Mask,sk.String.includes metadata in the outputs; for example,
meta={"colormap": "viridis"}controls the colormap used to display the image in Napari.
Summary#
Use
@sk.algorithmto convert a Python function into an executable algorithm.Annotate parameters using
parameters={}, type hints, default values, or variable names.Annotate return values with semantic types (
sk.Mask,sk.Image,sk.Float, etc.).Run algorithms interactively in Napari using
sk.to_napari()and passing it your algorithm.
Next steps#
In the next section, we will see how to further improve our algorithm by providing samples.